Predatory bacteria—bacteria that eat other bacteria—grow faster and consume more resources than non-predators in the same soil, according to a new study from Northern Arizona University. These active predators, which use wolfpack-like behavior, enzymes, and cytoskeletal “fangs” to hunt and feast on other bacteria, wield important power in determining where soil nutrients go. The results of the study, published this week in the journal mBio, show predation is an important dynamic in the wild microbial realm and suggest that these predators play an outsized role in how elements are stored in or released from soil.
Like every other life form on Earth, bacteria belong to intricate food webs in which organisms are connected to one another by what they consume and how. In macro webs, ecologists have long understood that when resources like grass and shrubs are added to lower levels of the web, predators at the top, such as wolves, often benefit. The research team, led by Bruce Hungate and other researchers from Northern Arizona University and Lawrence Livermore Laboratory, wanted to test whether the same was true in the microbial food webs found in wild soil.
“We’ve known predation plays a role in maintaining soil health, but we didn’t appreciate how significant predator bacteria are to these ecosystems before now,” said Hungate, who directs NAU’s Center for Ecosystem Science and Society.
To understand what and how much predator bacteria were consuming, the research team assembled a big picture using dozens of smaller data “snapshots:” 82 sets of data from 15 sites in a range of ecosystems. The team used information about how bacteria behave in culture to categorize bacteria as obligate or facultative predators. About 7 percent of all bacteria in the meta-analysis were identified as predators, and the majority of those were facultative, or omnivorous.
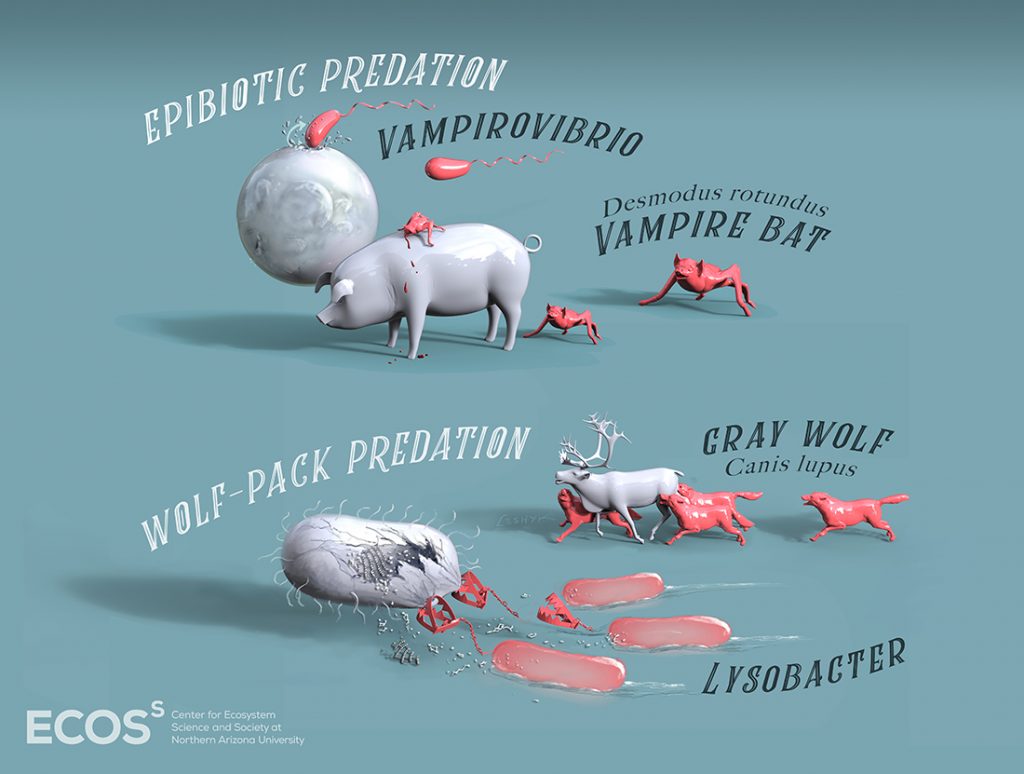
Obligate predatory bacteria like Bdellovibrionales and Vampirovibrionales grew 36 percent faster and took up carbon 211 percent faster than did non-predators. When the soil received a boost of carbon, predator bacteria used it to grow faster than other types. Researchers saw these effects in the omnivorous bacteria, as well, though the differences were less profound.
All the experiments were conducted using a state-of-the-art technique called quantitative Stable Isotope Probing, or qSIP. Researchers used labeled isotopes, which act a little like molecular hashtags, to track who is active and taking up nutrients in the soil. By sequencing the DNA in a soil sample and looking for these labels, the team could see who was growing and eating whom at the level of bacterial taxa.
“While analyzing my data, I noticed that Vampirovibrio was super enriched. Since we know Vampirovibrio is a predator, I became interested in looking for other potential predators in my other data,” said Brianna Finley, a postdoctoral research at University of California-Irvine and co-author on the study. “That we could pick up on these signals really validates qSIP as a tool.”
Soil ecosystems contain more carbon than is stored in all the plants on Earth, so understanding how carbon and other elements move among soil organisms is crucial to predicting future climate change. Because bacteria are so abundant in soil, they have an enormous role in how nutrients are stored or lost, and learning more about how predator bacteria act as “antibiotics” could have therapeutic implications down the road.
“Until now, predatory bacteria have not been a part of that soil story,” Hungate said. “But this study suggests that they are important characters who have a significant role determining the fate of carbon and other elements. These findings motivate us to take a deeper look at predation as a process.”
In addition to Hungate, the other NAU authors are Jane Marks, professor in the Department of Biological Sciences; Egbert Schwartz, professor in the Department of Biological Sciences; graduate research assistant Pete Chuckran, Paul Dijkstra, research professor in the Department of Biological Sciences; graduate student Megan Foley; Michaela Hayer, research associate for Ecoss; Ben Koch, senior research scientist for Ecoss; Michelle Mack, professor in the Department of Biological Sciences; Rebecca Mau, research associate, Pathogen & Microbiome Institute; Samantha Miller, research associate for Ecoss; Jeff Propster, research assistant for Ecoss; graduate research assistant Alicia Purcell; and former NAU researcher Bram Stone.
The research was supported by the Department of Energy’s Office of Biological and Environmental Research Genomic Sciences Program and a Lawrence Fellow award from the Lawrence Livermore National Laboratory.
Kate Petersen | Center for Ecosystem Science and Society