By Kerry Bennett
Office of the Vice President for Research
Ecological research focuses on understanding how population-level dynamics—such as the growth rate of a particular population of microbes—contribute to ecosystem-level processes. Ecosystem scientists researching climate change often study the role of microbes in the carbon cycle, for example, so knowing how quickly they grow is a fundamental metric to reaching that understanding.
Until now, however, scientists have not had the ability to measure growth rates of individual microbial populations other than by studying them in pure cultures within a petri dish. Unlike larger organisms such as birds, fish or mammals that can be tagged and monitored over time, individual microbial species are difficult to study effectively in their natural environment.
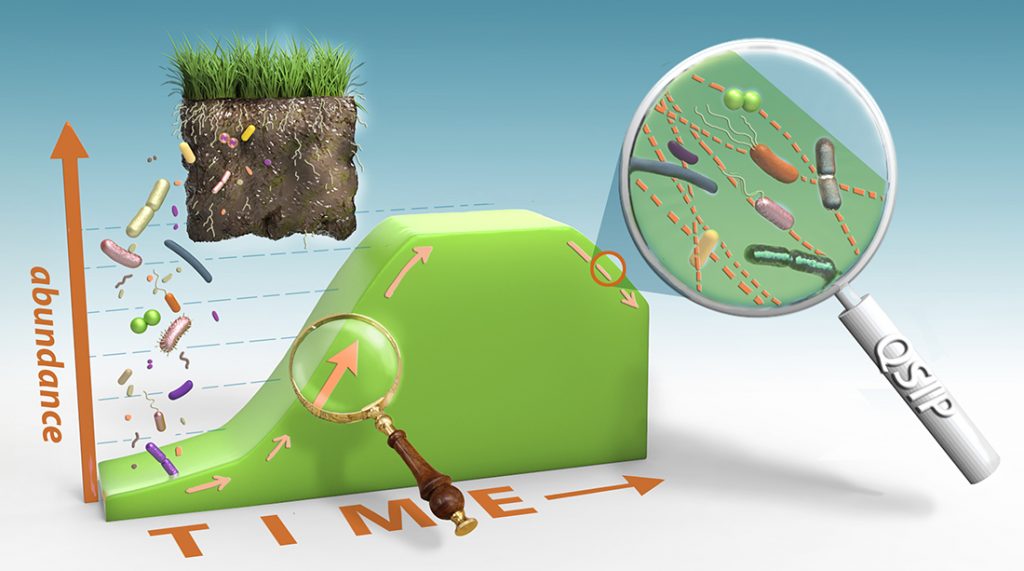
A team of scientists from Northern Arizona University’s Center for Ecosystem Science and Society (Ecoss) and Lawrence Livermore National Laboratory (LLNL) recently announced a major achievement in ecosystem science. Their research, published as “Estimating taxon-specific population dynamics in diverse microbial communities” in the journal Ecosphere, illustrates a powerful new technique to simultaneously measure the growth rates of hundreds of individual bacterial taxa in any given soil sample.
“Measuring the rate at which each microbe grows within an environmental sample is fundamental to understanding which organisms play the most important roles in natural and engineered environments that matter most to people, such as natural and agricultural soils, freshwaters and the human microbiome,” said lead author Ben Koch, senior research associate with Ecoss.
This project was a collaborative effort involving Steven Blazewicz and Jennifer Pett-Ridge of LLNL as well as nearly a dozen Ecoss faculty, laboratory staff, postdocs and students. Koch collaborated with associate research professor Paul Dijkstra, Regents’ professor and Ecoss director Bruce Hungate, professor Jane Marks and professor Egbert Schwartz, research associates Michaela Hayer and Rebecca Mau and former NAU postdocs Theresa McHugh, Ember Morrissey and Natasja van Gestel.
New technique based on technologies developed at NAU
This new measurement technique is based on the quantitative stable isotope probing (qSIP) technology Hungate developed. By adding rare stable isotopes to soil—tracers that contain heavy oxygen (18O)—and then sequencing the bacteria that incorporated that tracer, the scientists were able to measure microbial growth. The staff of NAU’s Environmental Genetics and Genomics Laboratory (EnGGen) provided the sequencing data, which was run through the QIIME pipeline developed by assistant professor Greg Caporaso, director of The Microbiome Center at NAU’s Pathogen and Microbiome Institute (PMI), to produce the final results.
“NAU’s strengths in this area have made it so easy with these types of analyses that require advanced bioinformatics,” Koch said
Koch and his team have shared the technique with other researchers at NAU, and it is already a major component of several projects recently awarded grants that are measuring the growth rates of organisms under varying conditions.
“Having the ability to measure the growth rates of individual microbial species in an environment that is close to their natural environment is a big step,” Koch said. “We can now take this basic model and apply it across a wide variety of situations—to study bacteria in the microbiome of a living animal, for example, or to study microorganisms in an industrial application like bioenergy production or even in a brewery. The sky’s the limit when we think about how we can apply this new technique.”
More about Ben Koch
Koch’s research explores how interactions among organisms regulate and are regulated by the cycling of energy and materials in ecosystems. He uses detailed natural history observations, experiments and quantitative modeling to study microbial ecology in a wide range of ecosystems, including streams, rivers, soils and the human microbiome.